Cancer Initiative and Genome Integrity
How does it start?
How does it progress?

How do we predict?

How do we treat?

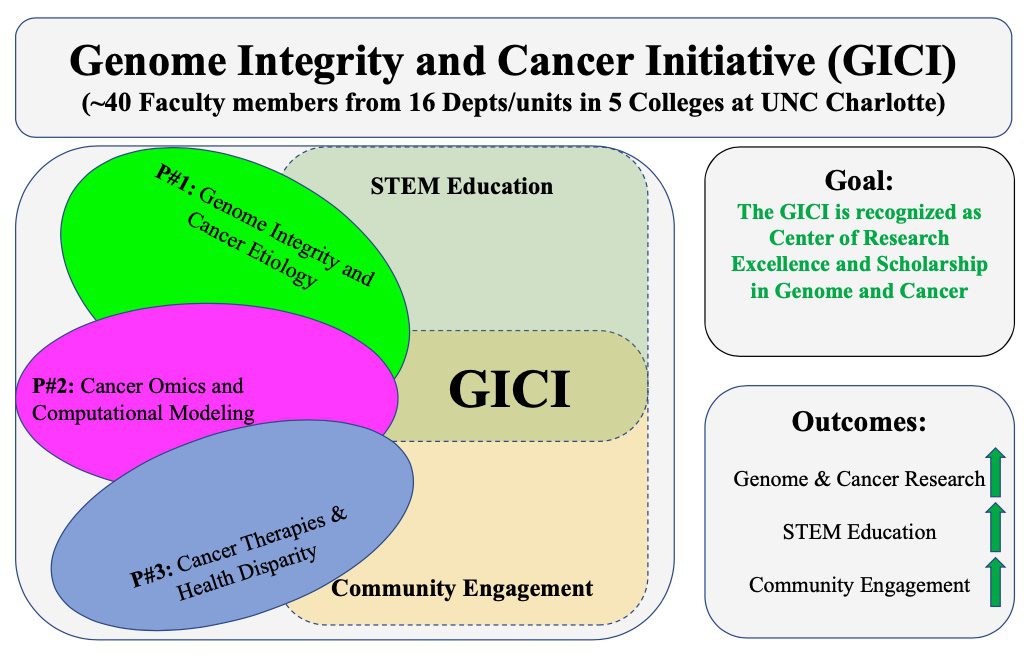
GICI: Genome Integrity and Cancer Initiative
The Genome Integrity and Cancer Initiative (GICI) is composed of three programs: (1) Genome integrity and cancer etiology (Lead- Shan Yan); (2) Cancer-omics and computational modeling (Co- Lead: Jun-tao Guo); (3) Cancer therapeutics and health disparity (Co-Lead: Didier Dréau) as well as STEM education and community engagement and outreach. The GICI will foster genome, omics, and cancer research, to support translational efforts to improve cancer treatment and community health, and promote STEM education and community engagement and outreach activities.
The GICI aims (1) to pursue excellence in cancer scholarship and research that address key fundamental cancer genetic, cellular and therapeutic challenges; (2) to pursue excellence in education by leveraging synergies between research and learning across our university community of scholars to offer undergraduate and graduate students both research experiences and innovative teaching; and (3) to promote community engagement and outreach activities in the local greater Charlotte region. Overall, GICI is fully committed to research excellence and scholarship as well as broad participation in both existing and emerging excellence. GICI is poised to make further strides in cancer research by harnessing the promises of (1) unity of the connecting participants (40 Faculty from 16 Depts/Units in 5 Colleges); (2) targeted individual and cancer (e.g., exercise, diet, genetic) factors; (3) cancer literacy and community awareness through outreach; and (4) strengthen the relationships with other regional, state, and national institutions. GICI will build on cutting-edge cancer research and embrace new pedagogy that fosters innovation in cancer research and education toward a recognized top-tier genome and cancer research center of excellence, through (1) the development and nurturing of a diverse intellectual research team that promotes cancer research investigation using molecular, cell, organism, model and community approaches; (2) the support of multi- and inter-disciplinary research opportunities/applications including those addressing cancer health disparities; and (3) the active participation in transforming STEM education and student involvement in cancer research and addressing cancer challenges at the local and national levels.
Upcoming Events
Dr. Shan Yan established the Exchange Group Charlotte Biology and Biotechnology with the North Carolina Biotechnology Center to bring the Distinguished Speaker in Genome Integrity Seminar series to campus. Upcoming dates:
- OCTOBER 8, 2021. Jianjun Zhao, PhD, Department of Cancer Biology, Lerner Research Institute, Cleveland Clinic. Cisplatin Upregulates Apurinic/Apyrimidinic Endonuclease 2 (APE2) Binding to Myosin Heavy-Chain 9 (MYH9), Provoking Mitochondrial Fragmentation and Acute Kidney Injury.
- NOVEMBER 5, 2021. Marcus Smolka, PhD, Department of Molecular Biology and Genetics, Cornell University. Boosting and Braking DNA Repair in Cancer.
Genome Integrity and Etiology Group (GIG)
GIG is a collaborative group of investigators involved in interdisciplinary research to understand DNA replication, recombination, damage and repair mechanisms in cancer and other human diseases. The GIG at UNC Charlotte aims to develop novel technologies and methodologies to investigate genome integrity. In addition to publishing high impact research, the GIG aims to provide an enriching training environment for Postdoctoral researchers, Graduate and Undergraduate students. Core members include leader Dr. Shan Yan (Molecular mechanisms of genome integrity and cancer etiology), Dr. Christine Richardson (DNA recombination and repair and cancer biology), Dr. Andrew Truman (Role of Molecular chaperones in the DNA damage response), Dr. Kausik Chakrabarti (RNA regulation in human pathogens and telomerase), and Dr. Junya Tomida (DNA repair, kinase regulation, cancer carcinogenesis and metastasis). This group meets every month for presentations by lab members, discussions and collaborations. Main Areas of Research in Program #1:
- Genome integrity, DNA repair, and DNA damage response
- DNA/RNA damage and cancer etiology
- Cellular signaling and molecular mechanisms
- Organismal, ecological and environmental analysis of genome integrity
Chakrabarti Lab
For more about Dr. Chakrabarti’s lab, visit his research lab webpage.
Research projects
- Structure and function of RNA and ribonucleoprotein complexes.
- Telomerase regulation and genome integrity in unicellular pathogens.
- Post-transcriptional mRNA processing in cell proliferation and cancer.
Representative Publications
- Dey A, Chakrabarti K. Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites. Int J Mol Sci. 2018;19(2). Epub 2018/01/25. doi: 10.3390/ijms19020333. PMID: 29364142;
- Ares M, Jr., Chakrabarti K*. Stuttering against marginotomy. Nat Struct Mol Biol. 2008;15(1):18-9. Epub 2008/01/08. doi: 10.1038/nsmb0108-18. PMID: 18176550.
- Sandhu R, Sanford S, Basu S, Park M, Pandya UM, Li B, Chakrabarti K*. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 2013;23(4):537-51. Epub 2013/03/13. doi: 10.1038/cr.2013.35. PubMed PMID: 23478302; PMCID: PMC3616428.
Dasgupta Lab
For more about Dr. Dasgupta’s lab, visit https://biology.charlotte.edu/people/dasgupta/
Research Projects
- Novel Targets in Cancer associated cachexia
- Dual Targets in Pancreatic Tumor and Cachectic Muscle
- Tumor-Host crosstalk
Representative Publications
- Dasgupta A, Schmitt RE, Kono T, Lee C,Zoberi MI, Epstein SA, Schneider JZ, Hernandez A, Grandgenett PM, Caffrey TC, DiMaio DJ, Hollingsworth MA, and Doles JD. Journal of
Experimental Medicine 2025 (in Press). Histidine decarboxylase inhibition attenuates
cancer-associated muscle wasting. *Joint co-corresponding authors. - Dasgupta A* ¥ , Gibbard DF*, Schmitt RE, Arneson-Wissink PC, Ducharme AM, Bruinsma
ES, Hawse JR, Jatoi A, Doles JD ¥ . PNAS 2023 Aug 22;120(34). A TGF-β/KLF10 signaling
axis regulates atrophy-associated genes to induce muscle wasting in pancreatic cancer.
*Joint co-first authors, ¥ Joint co-corresponding authors. - Dasgupta A, Arneson-Wissink PC, Schmitt RE, Cho DS, Ducharme AM, Hogenson TL,
Krueger EW, Bamlet WR, Zhang L, Razidlo G, Fernandez-Zapico ME, Doles JD. JCI Insight.
2022;7(2):e153842. Anti-cachectic regulator analysis reveals Perp-dependent anti-
tumorigenic properties of 3-methyladenine in pancreatic cancer. - Dasgupta A, Shukla SK, Vernucci E, RJ, Abrego J, Mulder SE,Mullen N, Graves G,
Buettner K, Thakur R, Murthy D, Attri KS, Chaika NV, Pacheco CG, Rai I, Engle DD,
Grandgenett PM, Punsoni M, Reames BN, Teoh-Fitzgerald M, Oberley-Deegan R, Yu F,
Klute KA, Hollingsworth MA, Zimmerman MC, Kamiya Mehla K, Sadoshima J, Tuveson D,
and Singh PK. Journal of Experimental Medicine. J Exp Med (2020) 217 (7). SIRT1-NOX4
Axis Regulates Muscle Atrophy in Cancer - Neyroud D, Laitano O, Dasgupta, A, Lopez C, Schmitt RE, Schneider JZ, Hammers DW,
Sweeney HL, Walter GA, Doles J, Judge SM, & Judge AR. (2023). Communications biology,
6(1), 519. Blocking muscle wasting via deletion of the muscle-specific E3 ligase MuRF1
impedes pancreatic tumor growth.
Dreau Lab
For more about Dr. Dreau’s lab, visit his research lab webpage
Research projects
- Chemokine heterodimerization and signaling
- Chemokine signaling and immunotherapy
- Inflammation and cancer progression
- 3D modeling of tumor and bone metastasis
Representative Publications
- Nguyen KTP, Volkman B, Dréau D, Nesmelova IV, 2022. A new obligate CXCL4-CXCL12
heterodimer for studying chemokine heterodimer activities and mechanisms. Sci Rep.
12(1):17204. doi: 10.1038/s41598-022-21651-0. PMID:36229490 - Nguyen KTP, Druhan LJ, Avalos BR, Zhai L, Rauova L, Nesmelova IV, Dréau D, 2020.
CXCL12-CXCL4 heterodimerization prevents CXCL12-driven breast cancer cell migration.
Cell Signal. 66:109488. doi: 10.1016/j.cellsig.2019.109488. PMID:31785332 - Rego SL, Helms RS, Dréau D, 2014. Breast tumor cell TACE-shed MCSF promotes pro-
angiogenic macrophages through NF-κB signaling. Angiogenesis. 17(3):573-85. doi:
10.1007/s10456-013-9405-2. PMID:24197832 - Lance A, Yang CC, Swamydas M, Dean D, Deitch S, Burg KJ, Dréau D, 2016. Increased
extracellular matrix density decreases MCF10A breast cell acinus formation in 3D culture
conditions. J Tissue Eng Regen Med. 10(1):71-80. doi: 10.1002/term.1675. PMID:23404906 - Bland E, Dréau D, Burg KJ, 2013. Overcoming hypoxia to improve tissue-engineering
approaches to regenerative medicine. J Tissue Eng Regen Med. 7(7):505-14. doi:
10.1002/term.540. PMID:22761177
Mukherjee Lab
For more about Dr. Mukherjee’s lab: https://biology.charlotte.edu/directory/pinku-mukherjee-phd/
and https://oncotab.com/about/
Research / Translational projects
- Immunotherapy of Epithelial Ductal Cancers (current focus: triple negative metastatic breast cancers and pancreatic ductal adenocarcinomas)
- Antibody engineering-Bispecific antibody/Bispecific T cell engagers
- CAR T cells and CAR NK cell therapies
- Theranoustics and ADCs (including nanoparticle loaded therapeutics)
- Immuno-Oncology related to tumor associated antigen, MUC1
- Metabolomics
- Oncogenic signaling
- Tumor cell Intrinsic Treatment Resistance Mechanisms
Representative Publications/Patents
- US9845362B2, Compositions comprising chimeric antigen receptors, T cells comprising the same, and methods of using the same
- US9090698B2, Tumor specific antibodies and uses therefor
- US20180043038A1, Tumor specific antibody conjugates and uses therefor
- Dahlia M. Besmer, Jennifer Curry, Lopamudra Das Roy, , et al and Pinku Mukherjee (2011). Pancreatic Ductal Adenocarcinoma (PDA) mice lacking Mucin 1 have a profound defect in tumor growth and metastasis. Cancer Research: 71:4432-4442. PMID: 21558393
- Sritama Nath and Pinku Mukherjee (2014) MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends in Molecular Medicine. 2014 Jun;20(6):332-42. PMID: 24667139
- Ru Zhou, Lopamudra Das Roy, Mahboubeh Yazdanifar, Chad Livasy and Pinku Mukherjee (2017). The use of tMUC1 highly specific chimeric antigen receptor-redirected T cells for the eradication of triple negative breast cancer, J Immunol May 1, 2017, 198 (1sup.) 198.10. PMID: 31178870
- Vanessa J Kelly, Shu-Ta Wu, et al, Pinku Mukherjee (2020). Preclinical evaluation of an 111In/225Ac theranostic targeting transformed MUC1 for triple negative breast cancer. Theranostics 2020; 10(15): 6946- 6958; doi:10.7150/thno.38236 PMID: 32550914
- Tarannum, Mubin; Hossain, Md; Holmes, Bryce; Yan, Shan; Mukherjee, Pinku; Vivero-Escoto, Juan. (2021). Advanced Nanoengineering Approach for Target-Specific, Spatiotemporal, and Ratiometric Delivery of Gemcitabine-Cisplatin Combination for Improved Therapeutic Outcome in Pancreatic Cancer. Small 2022 Jan;18(2):e2104449. PMID: 34758094
Richardson Lab
For more about Dr. Richardson’s lab, visit her research lab webpage.
Research projects
- DNA damage and translocations caused by bioflavonoids.
- DNA repair pathway choice.
- In vivo models of homologous recombination and non-homologous endjoining to repair double strand breaks.
Representative Publications
- Goodenow D, Emmanuel F, Berman C, Sahyouni M, Richardson C. Bioflavonoids cause DNA double-strand breaks and chromosomal translocations through topoisomerase II-dependent and -independent mechanisms. Mut Res: Gen Tox Env Gen. available online 2020.
- Bariar BB, Vestal CV, Deem B, Goodenow D, Ughetta M, Engledove W, Sahyouni M, Richardson C. Bioflavonoids promote stable translocations between MLL–AF9 breakpoint cluster regions independent of chromosomal context: model system to screen environmental risks. Env. Mol. Mutagenesis, 2019 PMID: 30387535.
- White, R, Sung, P, Vestal, CG, Benedetto, G., Cornelio, N., and Richardson, C. Double-strand break repair by interchromosomal recombination: an in vivo repair mechanism utilized by multiple somatic tissues in mammals. PlosONE, 8(12): 1-16, 2013. e84379. PMID: 24349572 PMCID: PMC3862804
Ren Lab
For more about Dr. Ren’s lab, visit his lab webpage
Research projects
- Exploring the Roles of Liquid-Liquid Phase Separation in Gene Regulation
- Single-Molecule Imaging of Epigenetic Processes
- Epigenetic Mechanisms in Cancers
- Therapeutic Targeting of Epigenetic Modifications and LLPS
Representative Publications
- Steven Ingersoll, Xiaojun Ren # (2025) Hypoxia-specific transcriptional condensates drive
metastasis. Trends Cell Biol. 2025 Jun;35(6):456-458. Pubmed/40379526 - Kyle Brown, Pin Yu Chew, Steven Ingersoll, Jorge R Espinosa, Anne Aguirre, Axel
Espinoza, Joey Wen, Kalkidan Astatike, Tatiana G Kutateladze, Rosana Collepardo-
Guevara, Xiaojun Ren # (2023) Principles of assembly and regulation of condensates of
Polycomb repressive complex 1 through phase separation. Cell Reports. 2023 Oct
31;42(10):113136. Pubmed/37756159 - Samantha Kent, Kyle Brown, Chou-Hsun Yang, Njood Alsaihati, Christina Tian, Haobin
Wang, Xiaojun Ren # (2020) Phase-Separated Transcriptional Condensates Accelerate
Target-Search Process Revealed by Live-Cell Single-Molecule Imaging. Cell Reports, 2020
Oct 13;33(2):108248. Pubmed/33053359 - Roubina Tatavosian, Samantha Kent, Kyle Brown, Tingting Yao, Huy Nguyen Duc, Thao
Ngoc Huynh, Chao Yu Zhen, Brian Ma, Haobin Wang, Xiaojun Ren # (2019) Nuclear
condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase
separation. J Biol Chem. 2019 Feb 1;294(5):1451-1463. Pubmed/30514760 - Roubina Tatavosian, Huy Nguyen Duc, Thao Ngoc Huynh, Dong Fang, Benjamin Schmitt,
Xiaodong Shi, Yiming Deng, Christopher Phiel, Tingting Yao, Zhiguo Zhang, Haobin Wang,
Xiaojun Ren # (2018) Live-cell single-molecule dynamics of PcG proteins imposed by the
DIPG H3. 3K27M mutation. Nat Commun. 2018 May 25;9(1):2080. Pubme/29802243
Tomida Lab
For more about Dr. Tomida’s lab, visit his research lab webpage.
Research projects
- Define factors leading to chemoresistance in prostate cancer (knockout mouse and cell
models). - Determine REV7 functions, which are critical for integrating multiple processes that
determine viability and genome stability. - Identify new roles for REV7 in the regulation of p53’s activities in cell cycle control,
apoptosis, DNA repair, and other cellular processes. - Characterize the protein complex of DNA repair proteins.
Representative Publications
- Tomida J*, Takata KI, Bhetawal S, Person MD, Chao HP, Tang DG, Wood RD. FAM35A
associates with REV7 and modulates DNA damage responses of normal and BRCA1-
defective cells. EMBO J. 2018;37(12). Epub 2018/05/24. DOI:
https://doi.org/10.15252/embj.201899543. PMID: 29789392. co-corresponding author - Fackrell K, Bobins L, Parul, and Tomida J*. FAM35A/SHLD2/RINN2: A novel
determinant of DSB repair pathway choice and genome stability in cancer. Environ Mol
Mutagen 61(7):709-715, 8/2020. e-Pub 7/2020. DOI: https://doi.org/10.1002/em.22379.
PMID: 32306447 - Biller M, Kabir S, Nipper S, Allen S, Kayali Y, Kuncik S, Sasanuma H, Wood R, Pei Zhou,
Vaziri C, Tomida J*. REV7 associates with ATRIP and inhibits ATR kinase activity.
bioRxiv. DOI: https://doi.org/10.1101/2025.01.17.633588. PMID: 39868202 - Biller M, Kabir S, Boado C, Nipper S, Saffa A, Tal A, Allen S, Sasanuma H, Dréau D,
Vaziri C, Tomida J*. REV7-p53 interaction inhibits ATM-mediated DNA damage
signaling. Cell Cycle 1-14, 4/2024. DOI:https://doi.org/10.1080/15384101.2024.2333227.
PMID: 38557443 - Tomida J, Takata K, Lange SS, Schibler AC, Yousefzadeh MJ, Bhetawal S, Dent SY,
Wood RD. REV7 is essential for DNA damage tolerance via two REV3L binding sites in
mammalian DNA polymerase ζ. Nucleic Acids Res 43(2):1000-11, 1/2015. e-Pub
1/2015. DOI: https://doi.org/10.1093/nar/gku1385. PMCID: PMC4333420
Truman Lab
For more about Dr. Truman’s lab, visit his research lab webpage.
Research projects
- Understanding the impact of Hsp70 post-translational modifications on the DNA damage
response - Characterizing the structure of the Ribonucleotide reductase (RNR)-chaperone megacomplex
using Cryo-EM and XL-MS - Identifying novel anticancer therapies that disrupt RNR-chaperone interactions
Representative Publications
- Omkar S†, Mitchem MM†, Hoskins JR, Shrader C†, Kline JT, Nitika†, Fornelli L, Wickner S,
Truman A.W.* (2024). Acetylation of the yeast Hsp40 chaperone protein Ydj1 fine-tunes
proteostasis and translational fidelity. PLOS Genetics. 2024;20(12):e1011338. doi:
10.1371/journal.pgen.1011338. - Omkar, S†., A. Rysbayeva‡, and Truman A.W.* (2023). Understanding chaperone
specificity: evidence for a ‘client code’. Trends Biochem Sci, 2023. 48(8): p. 662-664. - Nitika†, Zheng B, Ruan L, Kline JT, Omkar, S. †, Sikora J, Torres MT, Wang Y, Takakuwa
JE‡, Huguet R, Klemm C, Segarra VA, Winters MJ, Pryciak PM, Thorpe PH, Tatebayashi K,
Li R, Fornelli L and Truman A.W.* (2022). Comprehensive characterization of the Hsp70
interactome reveals novel client proteins and interactions mediated by post-translational
modifications. PLoS Biol 20(10): p. e3001839. - Knighton, L.E. †, Nitika†, Omkar, S. †, and Truman, A.W.* (2022). The C-terminal domain of
Hsp70 is responsible for paralog-specific regulation of ribonucleotide reductase. PLoS Genet
18, e1010079 - Nitika†, Blackman, J.S. ‡, Knighton, L.E. †, Takakuwa, L.E. ‡, Stuart K. Calderwood and
Truman A. W.* (2020) Chemogenomic screening identifies the Hsp70 co-chaperone DNAJA1
as a hub for anticancer drug resistance. Sci Rep 10, 13831. https://doi.org/10.1038/s41598-
020-70764-x.
van Oosten-Hawle Lab
For more about Dr. van Oosten-Hawle’s lab, visit her research lab webpage: https://www.vanoostenhawlelab.com/
Research projects
1. Transcellular chaperone signaling in the regulation of organismal health and disease
2. Gut-to-neuron stress signaling promoting neuroprotective and behavioral responses
3. Organismal proteostasis pathways in the regulation of the pathogen and DNA damage response
Representative Publications
1. Good SC, Dewison KM, Radford SE, van Oosten-Hawle P. Global Proteotoxicity Caused by Human β2 Microglobulin Variants Impairs the Unfolded Protein Response in C. elegans. International Journal of Molecular Sciences. 2021; 22(19):10752.
2. O’Brien, D., Jones, L., Good, S., Miles, J., Aston, R., Smith, C., Vijayabaskar, S., Westhead, D., and van Oosten-Hawle, P. (2018) A PQM-1-mediated response mediates transcellular chaperone signaling and organismal proteostasis. Cell Reports. 2018 Jun 26;23(13):3905-3919
3. Miles, J., van Oosten-Hawle P. Tissue-specific RNAi tools to identify components for systemic stress signaling. Journal of Visual Experiments (JOVE) (2020) May 16;(159).
4. van Oosten-Hawle, P., Porter, R.S., and Morimoto, R. I. (2013). Regulation of organismal proteostasis by transcellular chaperone signaling. Cell 153, 1366-1378.
Yan Lab
For more about Dr. Yan’s lab, visit his research lab webpage
Research projects
- To decipher the regulation and crosstalk of SSB/DSB repair and signaling pathways
- To elucidate the molecular mechanisms of nucleolar and mitochondrial DNA/RNA stability
- To dissect the molecular details of biomolecular condensates (liquid-liquid phase separation)
in genome integrity - To modulate the mechanisms of APE1, and APE2, and PARP1 in genome integrity for the
treatment of BRCA1-/BRCA2-related cancers
Representative Publications
- Zhao H, Li J, You Z, Lindsay HD, Yan S*. 2024. Distinct regulation of ATM signaling by DNA
single-strand breaks and APE1. Nature Communications. 15 (1): 6517. (PMCID:
PMC11306256; PMID: 39112456) DOI: https://doi.org/10.1038/s41467-024-50836-6 - Li J, Zhao H, McMahon A, Yan S*. 2022. APE1 assembles biomolecular condensates to
promote the ATR-Chk1 DNA damage response in nucleolus. Nucleic Acids Research.
50(18):10503-10525. (PMCID: PMC9561277; PMID: 36200829). DOI:
https://doi.org/10.1093/nar/gkac853 - Lin Y, Raj J, Li J, Ha A, Hossain MA, Richardson C, Mukherjee P, Yan S*. 2020. APE1
senses DNA single-strand breaks for repair and signaling. Nucleic Acids Research.
48(4):1925-1940. (PMCID: PMC7038996; PMID: 31828326) DOI:
https://doi.org/10.1093/nar/gkz1175 - Lin Y, Bai L, Cupello S, Hossain MA, Deem B, McLeod M, Raj J, Yan S*. 2018. APE2
promotes DNA damage response pathway from a single-strand break. Nucleic Acids
Research. 46 (5): 2479-2494. (PMCID: PMC5861430; PMID: 29361157) DOI:
https://doi.org/10.1093/nar/gky020 - Willis J, Patel Y, Lentz B , Yan S * . 2013. APE2 is required for ATR-Chk1 checkpoint
activation in response to oxidative stress. Proceedings of the National Academy of Sciences
of the United States of America. 110 (26): 10592-10597. (PMCID: PMC3696815; PMID:
23754435). DOI: http://dx.doi.org/10.1073/pnas.1301445110
Cancer-omics and computational modeling
With the decreasing costs of the next generation sequencing (NGS) approaches and the advancement of new technologies in the omics arena, a massive amount of cancer omics data have been generated and become available to the research community. For example, The Cancer Genome Atlas (TCGA) has sequenced over 20,000 primary cancer and corresponding normal samples covering 33 cancer types. These big data present a number of great challenges to the downstream analysis for cancer discovery, diagnosis, and treatment: how to efficiently integrate and visualize the data, what are the common and unique signatures or patterns among different cancer types, and how these data inform discovery of better and novel cancer therapeutics. Research areas synergize withthe Bioinformatics Resaerch Center and School of Data Science. Main Areas of Research in Program #2:
- Cancer transcriptomic and metabolomic research
- Data analytics and mathematical modeling
- Data integration and visualization
- Structural bioinformatics and functional studies
Cancer Therapeutics and Health Disparity
Targeted therapies have become the encouraging approaches to prevent cancer progression. However, disparity in tumors sensitivity and patient response and access to such treatments remains a challenge. Through investigations using relevant models including genetically engineered animals, immunocompetent mouse models, 3D cultures, the Cancer Therapeutics and Health Disparity program assesses innovative targeted therapies including immunotherapies, their delivery, diagnostic and monitoring. Moreover, measurement and interventions aim at reducing disparities in cancer treatment and survivorship modeled on efforts that limit diabetes and metabolic disease in the community will be extended to underrepresented cancer patient cohorts. Research areas synergize with the Center for Biomedical Engineering and Science (CBES). Main Areas of Research in Program #3:
- Tumor microenvironment and signaling
- Targeted therapy including immunotherapy
- Nanotechnology and targeted drug delivery
- Cancer disparity, cancer survivorship and community health access
STEM education and community engagement
The benefits of STEM education on the overall learning experience and academic success of undergraduate/graduate students are well documented. The proposed GICI will include the development of course-based research experiences (CUREs) including dissemination of research findings, research internships, and professional development modules. GICI also incorporates multiple STEM education initiatives and professional development programs. GICI faculty are actively engaged in STEM education and outreach with the community through various outreach programs including Discovery Place (public engagement) and HBCUs such as Johnson C. Smith University and NC A&T University (recruitment of graduate students). STEM education and community engagement activities developed by GICI are critical for establishing a large, diverse talent pool to meet the ever-increasing national demand for scientists in this field.