Immunology, Microbiology, and Virology
Augusto DG, Hollenbach JA. “HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection” Curr Opin Immunol. 2022 Jun;76:102178
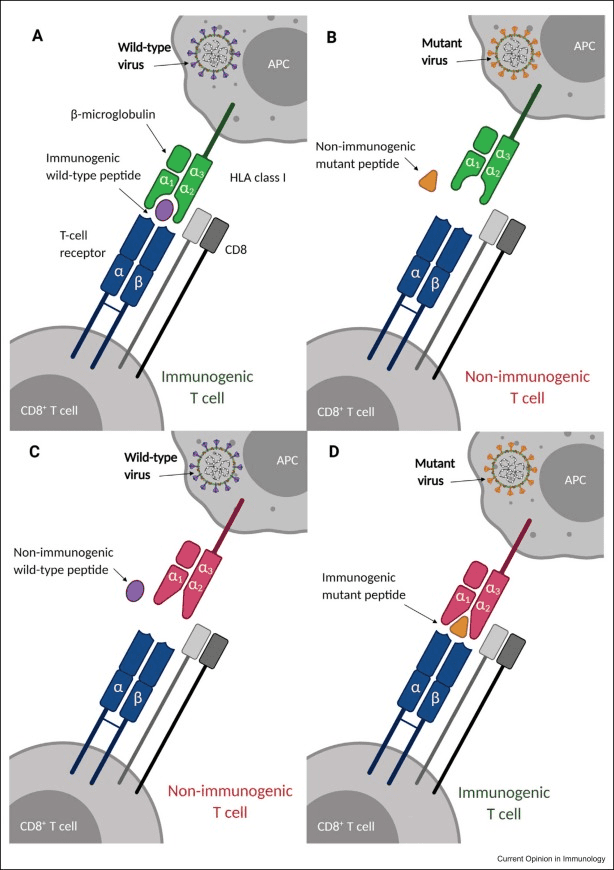
The Immunology, Microbiology, and Virology (IMV) Group encompasses a broad range of research and teaching activities related to the biology of “microbes” (viruses, bacteria, fungi, and protists), host-microbe interactions, microbial pathogenesis, the genetics and regulation of the host immune responses (innate and adaptive), and targeting the immune system as a strategy to treat cancer and other diseases. These studies at molecular, cellular, organismal, and population levels are central to our understanding of health, disease, and the environment, and for developing novel drugs, therapeutics, vaccines, and genetically modified organisms (e.g., crops) to prevent or/and treat infectious diseases, neurological conditions, or cancer.
Studies by IMV faculty are highly interdisciplinary, integrating biological and biochemical inquiry, genomics, public health, mathematics, and statistics. Several of these faculty are members of the Computational Intelligence to Predict Health and Environmental Risks (CIPHER) Center (focuses on genomics and computing technologies as applied to microbiology, biological and human diversity, and health), or have appointments with the School of Data Science that have interests in large data set analysis and the ethical and social considerations that come with it. These align with “Big Data” initiatives on campus.
Upcoming Events:
coming soon
Danillo Augusto – Immunogenetics, human population genetics, molecular basis of disease
mechanisms.
Tonya C. Bates – SEA-PHAGES Program (Science Education Alliance-Phage Hunters Advancing
Genomics and Evolutionary Science).
Sharon Bullock – SEA-PHAGES Program (Science Education Alliance-Phage Hunters Advancing
Genomics and Evolutionary Science).
Morgan Carter – bacterial-fungal symbioses and their impact on host health, plant-associated
microbes, molecular mechanisms of host-microbe interactions.
Kausik Chakrabarti – RNA-protein interactions regulating molecular functions related to cell
proliferation and genome integrity in blood-borne pathogens, Plasmodium falciparum (malaria)
and Trypanosoma brucei (a neurodegenerative disease), and in human cancers.
Richard Chi – Yeast Saccharomyces cerevisiae genetics, clathrin-mediated endocytosis,
endosome sorting and organelle genesis, autophagy.
Didier Dréau – Mechanisms of cancer metastasis, vascular and immune interactions during
cancer growth, inflammation, inflammasome studies, chemokine signaling with interest in
ligands’ functionality.
Farzana Ferdous – Immunological capacity of thrombocytes (nucleated platelets);
immunological properties of mesoporous silica nanoparticles.
Kristen Funk – Neuroinflammation of neuroinfectious and neurodegenerative diseases.
Valery Grdzelishvili – Molecular virology, virus-host interactions, anticancer therapy based on
replication-competent viruses (“oncolytic virotherapy”). Our lab mainly focuses on vesicular
stomatitis virus (VSV, family Rhabdoviridae) and other non-segmented negative strand (NNS)
RNA viruses (order Mononegavirales).
M. Brittany Johnson – Host-pathogen interactions and identification of therapeutic points of
intervention in meningitis and osteomyelitis.
Elaine Luo – Marine microbial ecology, environmental metagenomics, novel microbial diversity
in extreme/undersampled environments, and impacts of abundant viruses on microbial
diversity and biogeochemical (e.g., carbon) cycling.
Ian Marriott – Mechanisms of innate immunity in the brain, the role of the resident brain cells
in recognition of bacterial and viral pathogens, the role of the neuropeptide substance P in
microbe-induced inflammation.
Pinku Mukherjee – Cancer Biology and Immunology, assessing tumor-associated MUC1 as a
biomarker and therapeutic target for cancer.
Matthew W. Parrow – Eukaryotic microbiology, harmful algal blooms; applications of algae and
fungi in biotechnology.
Michelle B. Pass – SEA-PHAGES Program (Science Education Alliance-Phage Hunters Advancing
Genomics and Evolutionary Science).
Adam M. Reitzel – Evolution and ecology of coastal invertebrates, molecular biology of
circadian clocks, understanding the role of microbial communities in the development and
stress tolerance of marine species.
Andrew Truman – Understanding the role of molecular chaperones in cancer using quantitative
proteomics, molecular biology, systems biology, and model organisms (such as Saccharomyces
cerevisiae).
Bao-Hua (Alysa) Song – Plant evolutionary and ecological genetics and genomics, understanding
molecular mechanisms and evolution of complex trait variation significant in human health,
agriculture, and climate adaptation; dissecting novel genetic basis of broad resistance to
soybean cyst nematodes with single cell systems biology.
Sam Suptela – Mechanisms of innate immunity in the brain, the role of the resident cells in
recognition of bacterial and viral pathogens, the role of the neuropeptide substance P in
microbe-induced inflammation.